A Kalman Filter implementation in Rust
Table of Contents
I won’t detail how the Kalman Filter works as there are many great explanations out there (Bzarg, rlabbe, kalmanfilter.net). I am aware that there are Kalman filter implementations in Rust already, but it’s always fun to make something new, and I think this is a little bit different.
The repo is available at https://github.com/dw-labs-org/kfilter, the crate at https://crates.io/crates/kfilter/ and the documentation at https://docs.rs/kfilter/latest/kfilter/.
Version 1: Basic Implementation #
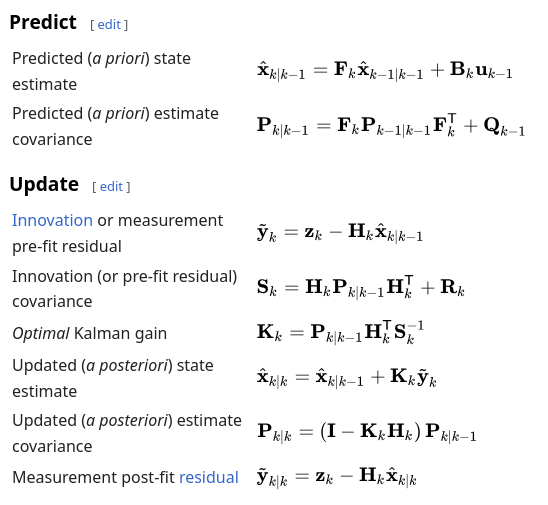
The notation I am using matches that on wikipedia, repeated below for convenience. The state vector x and each of the matrices are represented using the statically sized SMatrix from the excellent nalgebra crate for linear algebra.
I have used upper case variable names (F, P, Q, etc) to match the notation against rust convention, please forgive me, it helped to ensure the maths is correct.
In the code below, the Kalman struct contains the state and matrices and is generic over the matrix element type T (f32 or f64), the number of states N and the number of measurements M.
use nalgebra::{RealField, SMatrix, SVector};
/// Representation of Kalman filter
#[derive(Debug)]
struct Kalman<T, const N: usize, const M: usize> {
/// Estimated state
pub x: SVector<T, N>,
/// Discrete state transition
F: SMatrix<T, N, N>,
/// Covariance
pub P: SMatrix<T, N, N>,
/// Process Covariance
Q: SMatrix<T, N, N>,
/// Observation Matrix
H: SMatrix<T, M, N>,
/// Measurment covariance
R: SMatrix<T, M, M>,
}
The Kalman filter equations are split into predict and update. Predict estimates the state and covariance based on the model and its covariance. Update does the same using a measurement and its covariance.
impl<T: RealField + Copy, const N: usize, const M: usize> Kalman<T, N, M> {
pub fn predict(&mut self) {
self.x = self.F * self.x;
self.P = self.F * self.P * self.F.transpose() + self.Q;
}
pub fn update(&mut self, z: &SVector<T, M>) {
// innovation
let y = z - (self.H * self.x);
// innovation covariance
let S = self.H * self.P * self.H.transpose() + self.R;
// Optimal gain
let K = self.P * self.H.transpose() * S.try_inverse().unwrap();
// state update
self.x += K * y;
// covariance update
self.P = (SMatrix::identity() - K * self.H) * self.P;
}
pub fn new(
F: SMatrix<T, N, N>,
Q: SMatrix<T, N, N>,
H: SMatrix<T, M, N>,
R: SMatrix<T, M, M>,
) -> Self {
Kalman {
x: SMatrix::zeros(),
F,
P: SMatrix::zeros(),
Q,
H,
R,
}
}
}
Below is example usage with a 1D constant velocity model and noisy position measurments.
fn const_velocity() {
// Example usage with position and velocity states and measured position
// Model assumes constant velocity
// Timestep
let dt = 0.1;
let F = Matrix2::new(1.0, 0.1, 0.0, 1.0);
let Q = Matrix2::identity();
let H = Matrix1x2::new(1.0, 0.0);
let R = Matrix1::identity() * 0.5;
let mut kalman = Kalman::new(F, Q, H, R);
// Create a random number generator (using rand_distr crate)
let position_error = Normal::new(0.0, 1.0).unwrap();
let mut rng = rand::thread_rng();
// set initial state and covariance
kalman.x = Vector2::new(position_error.sample(&mut rng), 0.0);
kalman.P = Q;
// Record state evolution
const T: usize = 40;
const VELOCITY: f64 = 1.0;
let positions = (0..T).map(|t| (t as f64) * VELOCITY).collect::<Vec<f64>>();
let mut measured = Vec::with_capacity(T);
let mut predicted = Vec::with_capacity(T);
let mut updated = Vec::with_capacity(T);
let mut covariance = Vec::with_capacity(T);
// iterate though time in seconds
for position in &positions {
// The noise corrupted measurement
let measurement = position + position_error.sample(&mut rng);
measured.push(measurement);
// kalman predict and update
kalman.predict();
predicted.push(kalman.x.x);
kalman.update(&Matrix1::new(measurement));
updated.push(kalman.x.x);
covariance.push(kalman.P.m11);
}
}
The recorded real, measured, predicted and updated positions look like this (plotted using plotters):
See version1 for the full code including plotting the graph.
This is probably enough for a lot of projects, but I would like to take it further. There are some problems that need addressing…
Issues #
- Some systems have inputs which modify their state via a control matrix.
- The EKF requires a non-linear state transition function for estimating
xand the covariance is updated using the Jacobian of this function forF. - Even linear systems may have varying process and measurement covariances, which are currently immutable.
- The measurement vector does not have to be a fixed size. The KF is often used for sensor fusion and each sensor could have different noise and observation matrices (including a different shape, which doesn’t work with the current const generics)
H.transpose()gets called twice inupdate()- an easy optimisation.
Version 2: Adding Systems #
The aim here is to address issues 1, 2 and 3 by making the system that the KF operates on generic. The system has 2 roles to fulfill:
- Transition from the current state to the next
- Generate a transition matrix for updating the covariance
For this to work, the system is required to implement the following trait. The new const generic U is the size of the input vector.
pub trait System<T, const N: usize, const U: usize> {
/// transition to the next state, returning a reference to it
fn step(&mut self) -> &SVector<T, N>;
/// transition to the next state with an input, returning a reference to it
fn step_with_input(&mut self, u: SVector<T, U>) -> &SVector<T, N>;
/// Get a reference to the transition matrix (Jacobian)
fn transition(&self) -> &SMatrix<T, N, N>;
/// Get a reference to the process covariance matrix
fn covariance(&self) -> &SMatrix<T, N, N>;
/// Get a reference to the state
fn state(&self) -> &SVector<T, N>;
/// Get a mutable reference to the state
fn state_mut(&mut self) -> &mut SVector<T, N>;
}
And some changes are made to the KF to use a type that implements System.
/// Representation of Kalman filter
#[derive(Debug)]
struct Kalman<T, const N: usize, const M: usize, const U: usize, S: System<T, N, U>> {
/// Covariance
pub P: SMatrix<T, N, N>,
/// The associated system
pub system: S,
/// Observation Matrix
H: SMatrix<T, M, N>,
/// Measurment covariance
R: SMatrix<T, M, M>,
}
impl<T: RealField + Copy, const N: usize, const M: usize, const U: usize, S: System<T, N, U>>
Kalman<T, N, M, U, S>
{
pub fn predict(&mut self) {
self.system.step();
let F = self.system.transition();
self.P = F * self.P * F.transpose() + self.system.covariance();
}
pub fn predict_with_input(&mut self, u: SVector<T, U>) {
self.system.step_with_input(u);
let F = self.system.transition();
self.P = F * self.P * F.transpose() + self.system.covariance();
}
pub fn update(&mut self, z: &SVector<T, M>) {
// innovation
let y = z - (self.H * self.system.state());
// innovation covariance
let S = self.H * self.P * self.H.transpose() + self.R;
// Optimal gain
let K = self.P * self.H.transpose() * S.try_inverse().unwrap();
// state update
*self.system.state_mut() += K * y;
// covariance update
self.P = (SMatrix::identity() - K * self.H) * self.P;
}
pub fn new(system: S, H: SMatrix<T, M, N>, R: SMatrix<T, M, M>) -> Self {
Kalman {
P: SMatrix::zeros(),
system,
H,
R,
}
}
}A linear system that implements System:
pub struct LinearSystem<T, const N: usize, const U: usize> {
x: SVector<T, N>,
F: SMatrix<T, N, N>,
Q: SMatrix<T, N, N>,
B: SMatrix<T, N, U>,
}
impl<T: RealField + Copy, const N: usize, const U: usize> LinearSystem<T, N, U> {
pub fn new(F: SMatrix<T, N, N>, Q: SMatrix<T, N, N>, B: SMatrix<T, N, U>) -> Self {
LinearSystem {
x: SMatrix::zeros(),
F,
Q,
B,
}
}
}
impl<T: RealField + Copy, const N: usize> LinearSystem<T, N, 0> {
pub fn new_no_input(F: SMatrix<T, N, N>, Q: SMatrix<T, N, N>) -> Self {
LinearSystem {
x: SMatrix::zeros(),
F,
Q,
B: SMatrix::zeros(),
}
}
}
impl<T: RealField + Copy, const N: usize, const U: usize> System<T, N, U>
for LinearSystem<T, N, U>
{
fn step(&mut self) -> &SVector<T, N> {
self.x = self.F * self.x;
&self.x
}
fn step_with_input(&mut self, u: SVector<T, U>) -> &SVector<T, N> {
self.x = self.F * self.x + self.B * u;
&self.x
}
fn transition(&self) -> &SMatrix<T, N, N> {
&self.F
}
fn covariance(&self) -> &SMatrix<T, N, N> {
&self.Q
}
fn state(&self) -> &SVector<T, N> {
&self.x
}
fn state_mut(&mut self) -> &mut SVector<T, N> {
&mut self.x
}
}
This feels a little bit overcomplicated and convoluted, however it does allow for systems with inputs to be modelled and indeed any non-linear state transition, provided the Jacobian is calculable and the System trait is implemented.
Type aliases can make it a bit more pleasant to use. Getting back to the simplicity of the first implementation:
type LinearSystemNoInput<T, const N: usize> = LinearSystem<T, N, 0>;
type KalmanLinearNoInput<T, const N: usize, const M: usize> =
Kalman<T, N, M, 0, LinearSystemNoInput<T, N>>;
impl<T: RealField + Copy, const N: usize, const M: usize> KalmanLinearNoInput<T, N, M> {
pub fn new_linear_no_input(
F: SMatrix<T, N, N>,
Q: SMatrix<T, N, N>,
H: SMatrix<T, M, N>,
R: SMatrix<T, M, M>,
) -> Self {
Kalman::new(LinearSystemNoInput::new(F, Q, SMatrix::zeros()), H, R)
}
}
And the predict and update functions can be used as before.
So that is 1, 2 and part of 3 sorted. Now to look at the measurement and update stage of the KF. The source code can found here.
Version 3: Measurements #
The problem outlined in Issue 4 stems from the fact that the const generic M is fixed for a specific instance of the Kalman struct. Therefore the update function needs to be modified to accept any size measurement, along with its corresponding observation and covariance matrices H and R.
There are probably a few ways to do this and I certainly tried a lot that didn’t work. Ultimately I settled on using a KalmanUpdate trait to specify the required behaviour of the KF when updating with a Measurement struct.
trait KalmanUpdate<T, const N: usize, const M: usize> {
fn update(&mut self, measurement: &Measurement<T, N, M>);
}
impl<T: RealField + Copy, const N: usize, const M: usize, const U: usize, S: System<T, N, U>>
KalmanUpdate<T, N, M> for Kalman<T, N, U, S>
{
fn update(&mut self, measurement: &Measurement<T, N, M>) {
let H_transpose = measurement.H.transpose();
// innovation
let y = measurement.z - (measurement.H * self.system.state());
// innovation covariance
let S = measurement.H * self.P * H_transpose + measurement.R;
// Optimal gain
let K = self.P * H_transpose * S.try_inverse().unwrap();
// state update
*self.system.state_mut() += K * y;
// covariance update
self.P = (SMatrix::identity() - K * measurement.H) * self.P;
}
}
pub struct Measurement<T, const N: usize, const M: usize> {
pub z: SVector<T, M>,
pub H: SMatrix<T, M, N>,
pub R: SMatrix<T, M, M>,
}
The Kalman struct is no longer constrained by the measurement vector length M and measurements can vary in length, fixing issue 4 and the varying measurement covariance in issue 3. Issue 5 has also been fixed by calling the H.transpose() function just once. There are still some problems however…
For a proper EKF implementation, the map from state space to observation space performed by the
Hmatrix can be non-linear. Similar to theFmatrix in the system model, there should be a function to perform the mapping and another to return the Jacobian.For a simple linear KF with constant transition/observation/covariances, this is now more complicated to use as a
Measurementinstance needs to be kept around or created each timestep. The only way around this is probably a wrapper aroundKalmanfor single measurement systems.The transition and observation matrix transposes are still calculated every time-step which is unnecessary for linear systems
Source code: version3.
Version 4: EKF #
A nonlinear observation means creating a trait for measurements analogous to the System trait in Version 2.
pub trait Measurement<T, const N: usize, const M: usize> {
/// Calculate the innovation (y) based on the measurement and the observation mapping
fn innovation(&self, x: &SVector<T, N>) -> SVector<T, M>;
/// Get the measurement covariance
fn covariance(&self) -> &SMatrix<T, M, M>;
/// Get the observation matrix
fn observation(&self) -> &SMatrix<T, M, N>;
}
pub struct LinearMeasurement<T, const N: usize, const M: usize> {
pub z: SVector<T, M>,
pub H: SMatrix<T, M, N>,
pub R: SMatrix<T, M, M>,
}
impl<T: RealField + Copy, const N: usize, const M: usize> Measurement<T, N, M>
for LinearMeasurement<T, N, M>
{
fn innovation(&self, x: &SVector<T, N>) -> SVector<T, M> {
self.z - (self.H * x)
}
fn covariance(&self) -> &SMatrix<T, M, M> {
&self.R
}
fn observation(&self) -> &SMatrix<T, M, N> {
&self.H
}
}
And the Kalman struct and KalmanUpdate trait are modified accordingly:
trait KalmanUpdate<T, const N: usize, const M: usize, ME: Measurement<T, N, M>> {
fn update(&mut self, measurement: &ME);
}
impl<
T: RealField + Copy,
const N: usize,
const M: usize,
const U: usize,
S: System<T, N, U>,
ME: Measurement<T, N, M>,
> KalmanUpdate<T, N, M, ME> for Kalman<T, N, U, S>
{
fn update(&mut self, measurement: &ME) {
let H_transpose = measurement.observation().transpose();
// innovation
let y = measurement.innovation(self.system.state());
// innovation covariance
let S = measurement.observation() * self.P * H_transpose + measurement.covariance();
// Optimal gain
let K = self.P * H_transpose * S.try_inverse().unwrap();
// state update
*self.system.state_mut() += K * y;
// covariance update
self.P = (SMatrix::identity() - K * measurement.observation()) * self.P;
}
}Source code: version4.
Version 5: Back to basics #
With all the additional features to make the KF and EKF functionality as configurable as possible, creating a basic fixed linear KF is now quite complicated compared to Version 1. To make this easier, Kalman1M wraps the Kalman struct, storing the measurement matrices.
/// Kalman filter with a fixed shape measurement
struct Kalman1M<
T,
const N: usize,
const U: usize,
const M: usize,
S: System<T, N, U>,
ME: Measurement<T, N, M>,
> {
kalman: Kalman<T, N, U, S>,
measurement: ME,
}
impl<T: RealField + Copy, const N: usize, const M: usize>
Kalman1M<T, N, 0, M, LinearSystemNoInput<T, N>, LinearMeasurement<T, N, M>>
{
pub fn new_linear_no_input(
F: SMatrix<T, N, N>,
Q: SMatrix<T, N, N>,
H: SMatrix<T, M, N>,
R: SMatrix<T, M, M>,
) -> Self {
Kalman1M {
kalman: Kalman::new_linear_no_input(F, Q),
measurement: LinearMeasurement {
z: SMatrix::zeros(),
H,
R,
},
}
}
pub fn predict(&mut self) {
self.kalman.predict();
}
pub fn update(&mut self, z: SVector<T, M>) {
self.measurement.z = z;
self.kalman.update(&self.measurement);
}
}
This looks far from basic but the end result hopefully is. Kalman1M can be constructed with the 4 matrices and predict() and update() can be used as before.
Source code: version5.
Version 6: Performance #
The aim here is to remove the unnecessary transpose calculations that are performed at every timestep, which for a linear system do not change. Adding an extra function the System trait:
pub trait System<T, const N: usize, const U: usize> {
...
/// Get the transpose of the transition matrix
fn transition_transpose(&self) -> &SMatrix<T, N, N>;
...
}
Then for the LinearSystem, the transpose can be recorded on creation:
impl<T: RealField + Copy, const N: usize, const U: usize> LinearSystem<T, N, U> {
pub fn new(F: SMatrix<T, N, N>, Q: SMatrix<T, N, N>, B: SMatrix<T, N, U>) -> Self {
LinearSystem {
x: SMatrix::zeros(),
F,
F_t: F.transpose(),
Q,
B,
}
}
}
And be retrieved each timestep:
impl<T: RealField + Copy, const N: usize, const U: usize> System<T, N, U>
for LinearSystem<T, N, U>
{
...
fn transition_transpose(&self) -> &SMatrix<T, N, N> {
&self.F_t
}
...
}
impl<T: RealField + Copy, const N: usize, const U: usize, S: System<T, N, U>> Kalman<T, N, U, S> {
pub fn predict(&mut self) {
self.system.step();
self.P = self.system.transition() * self.P * self.system.transition_transpose()
+ self.system.covariance();
}
}
The same changes can be made to the Measurement trait to store the transpose of the observation matrix and return a reference to it at each update() call.
Source code: version6.
Version 7: Tidying up with traits #
The current implementation has one more issue I would like to address. The System trait contains both the step and step_with_input functions, which should be mutually exclusive as no type should implement both. It also means both predict and predict_with_input can be called on Kalman which does’t make sense and could lead to errors.
The step functionality is move into 2 new traits, InputSystem and NoInputSystem:
pub trait InputSystem<T, const N: usize, const U: usize>: System<T, N, U> {
/// transition to the next state with input,
/// returning a reference to it
fn step(&mut self, u: SVector<T, U>) -> &SVector<T, N>;
}
pub trait NoInputSystem<T, const N: usize>: System<T, N, 0> {
/// transition to the next state, returning a reference to it
fn step(&mut self) -> &SVector<T, N>;
}
Then systems can implement these according to whether they can take inputs or not.
Then to prevent both predict functions being available, 2 new traits are created for Kalman:
trait KalmanPredict<T, const N: usize> {
/// predict the next state and return it. Also update covariance
fn predict(&mut self) -> &SVector<T, N>;
}
trait KalmanPredictInput<T, const N: usize, const U: usize> {
/// predict the next state with an input vector and return it. Also update covariance
fn predict(&mut self, u: SVector<T, U>) -> &SVector<T, N>;
}
And Kalman only implements each trait when its associated system implements the corresponding system trait, as shown below:
/// Implement the predict stage for a system with no input
impl<T: RealField + Copy, const N: usize, S: NoInputSystem<T, N>> KalmanPredict<T, N>
for Kalman<T, N, 0, S>
{
fn predict(&mut self) -> &SVector<T, N> {
self.system.step();
self.P = self.system.transition() * self.P * self.system.transition_transpose()
+ self.system.covariance();
self.system.state()
}
}
/// Implement the predict stage for a system with input
impl<T: RealField + Copy, const N: usize, const U: usize, S: InputSystem<T, N, U>>
KalmanPredictInput<T, N, U> for Kalman<T, N, U, S>
{
fn predict(&mut self, u: SVector<T, U>) -> &SVector<T, N> {
self.system.step(u);
self.P = self.system.transition() * self.P * self.system.transition_transpose()
+ self.system.covariance();
self.system.state()
}
}
And I think that is enough messing with the library. The source code up to this point can be found at version7.
Nonlinear system #
As a final addition, this is the generic non-linear system in the library. By supplying a step_fn that implements f(x,u) as well as returning the jacobian and process covariance, an EKF
pub struct NonLinearSystem<T, const N: usize, const U: usize> {
/// System state
x: SVector<T, N>,
/// Process Covariance, updated after step_fn() call.
Q: SMatrix<T, N, N>,
/// Jacobian, updated after jacobian() call.
F: SMatrix<T, N, N>,
/// Jacobian transpose, updated after jacobian() call.
F_t: SMatrix<T, N, N>,
/// Function that steps from current state to next with an input
/// Returns the new state, the jacobian and the process covariance
step_fn: StepInput<T, N, U>,
}
impl<T: RealField + Copy, const N: usize, const U: usize> InputSystem<T, N, U>
for NonLinearSystem<T, N, U>
{
fn step(&mut self, u: SVector<T, U>) -> &SVector<T, N> {
// Get updated state, jacobian and process covariance
let r = (self.step_fn)(self.x, u);
self.x = r.state;
self.F = r.jacobian;
self.F_t = self.F.transpose();
self.Q = r.covariance;
// Return state
self.state()
}
}
Conclusion and future work #
I am pretty happy with how things have worked out and I believe it meets to goal of a flexible library for both the KF and EKF.
Before publishing to crates.io I also added different constructors for common configurations, a nicer way of setting initial state and covariance values, some documentation and examples.
I would like to look at methods for ensuring the covariance remains symmetric as this can sometimes be necessary due to numerical issues. There are also other forms of the KF like the unscented Kalman filter that I would like to learn more about and implement.